Bacterial taxonomy Bacterial taxonomy is subfield of taxonomy devoted to the classification of bacteria specimens into taxonomic ranks. Archaeal taxonomy are governed by the same rules. In the scientific classification established by Carl Linnaeus,[1] each species is assigned to a genus resulting in a two-part name. This name denotes the two lowest levels in a hierarchy of ranks, increasingly larger groupings of species based on common traits. Of these ranks, domains are the most general level of categorization. Presently, scientists classify all life into just three domains, Eukaryotes, Bacteria and Archaea.[2] Bacterial taxonomy is the classification of strains within the domain Bacteria into hierarchies of similarity. This classification is similar to that of plants, mammals, and other taxonomies. However, biologists specializing in different areas have developed differing taxonomic conventions over time. For example, bacterial taxonomists name types based on descriptions of strains. Zoologists among others use a type specimen instead. DiversityBacteria (prokaryotes, together with Archaea) share many common features. These commonalities include the lack of a nuclear membrane, unicellularity, division by binary-fission and generally small size. The various species can be differentiated through the comparison of several characteristics, allowing their identification and classification. Examples include:
HistoryFirst descriptionsBacteria were first observed by Antonie van Leeuwenhoek in 1676, using a single-lens microscope of his own design.[3] He did not distinguish bacteria as a separate type of microorganism, calling all microorganisms, including bacteria, protists, and microscopic animals, "animalcules". He published his observations in a series of letters to the Royal Society.[4][5][6] Early described genera of bacteria include Vibrio and Monas, by O. F. Müller (1773, 1786), then classified as Infusoria (however, many species before included in those genera are regarded today as protists, which are eukaryotes); Polyangium, by H. F. Link (1809), the first bacterium still recognized today; Serratia, by Bizio (1823); and Spirillum, Spirochaeta and Bacterium, by Ehrenberg (1838).[7][8] The term Bacterium, introduced as a genus by Ehrenberg in 1838,[9] became a catch-all for rod-shaped cells.[7] Early formal classifications In 1857, bacteria were classified as plants constituting the class Schizomycetes, which along with the Schizophyceae (blue green algae/Cyanobacteria) formed the phylum Schizophyta.[11] Haeckel in 1866 placed the group in the phylum Moneres (from μονήρης: simple) in the kingdom Protista and defines them as completely structureless and homogeneous organisms, consisting only of a piece of plasma.[10] He subdivided the phylum into two groups:[10]
The classification of Ferdinand Cohn (1872) was influential in the nineteenth century, and recognized six genera: Micrococcus, Bacterium, Bacillus, Vibrio, Spirillum, and Spirochaeta.[7] The group was later reclassified as the Prokaryotes by Chatton in 1925.[16] The classification of Cyanobacteria (colloquially "blue green algae") has been fought between being algae or bacteria (for example, Haeckel classified Nostoc in the phylum Archephyta of Algae[10]). in 1905, Erwin F. Smith accepted 33 valid different names of bacterial genera and over 150 invalid names,[17] and Vuillemin, in a 1913 study,[18] concluded that all species of the Bacteria should fall into the genera Planococcus, Streptococcus, Klebsiella, Merista, Planomerista, Neisseria, Sarcina, Planosarcina, Metabacterium, Clostridium, Serratia, Bacterium, and Spirillum. in 1875, Cohn[19] recognized four tribes: Spherobacteria, Microbacteria, Desmobacteria, and Spirobacteria. Stanier and van Neil in 1941[20] recognized the kingdom Monera with two phyla, Myxophyta and Schizomycetae, the latter comprising classes Eubacteriae (three orders), Myxobacteriae (one order), and Spirochetae (one order). In 1962, Bisset[21] distinguished 1 class and 4 orders: Eubacteriales, Actinomycetales, Streptomycetales, and Flexibacteriales. Walter Migula's system (1897),[22] which was the most widely accepted system of its time and included all then-known species but was based only on morphology, contained the three basic groups Coccaceae, Bacillaceae, and Spirillaceae, but also Trichobacterinae for filamentous bacteria. Orla-Jensen in 1909[23] established two orders: Cephalotrichinae (seven families) and Peritrichinae (presumably with only one family). Bergey et al. in 1925[24] presented a classification which generally followed the 1920 Final Report of the Society of American Bacteriologists Committee (Winslow et al.), which divided class Schizomycetes into four orders: Myxobacteriales, Thiobacteriales, Chlamydobacteriales, and Eubacteriales, with a fifth group being four genera considered intermediate between bacteria and protozoans: Spirocheta, Cristospira, Saprospira, and Treponema. However, different authors often reclassified the genera due to the lack of visible traits to go by, resulting in a poor state which was summarised in 1915 by Robert Earle Buchanan.[25] By then, the whole group received different ranks and names by different authors, namely:
Furthermore, the families into which the class was subdivided changed from author to author and for some, such as Zipf (1917), the names were in German and not in Latin.[29] The first edition of the Bacteriological Code in 1947 set a standardised system and authority for the classification of Bacteria.[30] A. R. Prévot's system (1958)[31][32] had four subphyla and eight classes, as follows:
Informal groups based on Gram stainingDespite there being little agreement on the major subgroups of the Bacteria, Gram staining results were most commonly used as a classification tool. Consequently, until the advent of molecular phylogeny, the Kingdom Prokaryota was divided into four divisions,[41] A classification scheme still formally followed by Bergey's manual of systematic bacteriology for tome order[42]
Molecular era"Archaic bacteria" and Woese's reclassification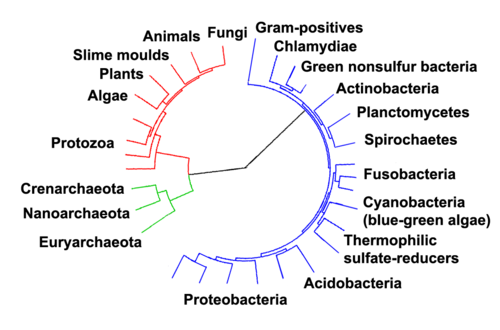 Woese argued that the bacteria, archaea, and eukaryotes represent separate lines of descent that diverged early on from an ancestral colony of organisms.[45][46] However, a few biologists argue that the Archaea and Eukaryota arose from a group of bacteria.[47] In any case, it is thought that viruses and archaea began relationships approximately two billion years ago, and that co-evolution may have been occurring between members of these groups.[48] It is possible that the last common ancestor of the bacteria and archaea was a thermophile, which raises the possibility that lower temperatures are "extreme environments" in archaeal terms, and organisms that live in cooler environments appeared only later.[49] Since the Archaea and Bacteria are no more related to each other than they are to eukaryotes, the term prokaryote's only surviving meaning is "not a eukaryote", limiting its value.[50] With improved methodologies it became clear that the methanogenic bacteria were profoundly different and were (erroneously) believed to be relics of ancient bacteria[51] thus Carl Woese, regarded as the forerunner of the molecular phylogeny revolution, identified three primary lines of descent: the Archaebacteria, the Eubacteria, and the Urkaryotes, the latter now represented by the nucleocytoplasmic component of the Eukaryotes.[52] These lineages were formalised into the rank Domain (regio in Latin) which divided Life into 3 domains: the Eukaryota, the Archaea and the Bacteria.[2] In 2023, the Prokaryotic Code added the ranks of domain and kingdom to the prokaryotic nomenclature. The names of Bacteria and Archaea are validly-published taxa following Oren and Goker's publication that use these new rules.[53] SubdivisionsIn 1987 Carl Woese divided the Eubacteria into 11 divisions based on 16S ribosomal RNA (SSU) sequences, which with several additions are still used today.[54][55] Oren and Goker has also validly published a number of kingdoms as a layer higher than the division/phylum:[53]
OppositionWhile the three domain system is widely accepted,[56] some authors have opposed it for various reasons. One prominent scientist who opposed the three domain system was Thomas Cavalier-Smith, who proposed that the Archaea and the Eukaryotes (the Neomura) stem from Gram positive bacteria (Posibacteria), which in turn derive from gram negative bacteria (Negibacteria) based on several logical arguments,[57][58] which are highly controversial and generally disregarded by the molecular biology community (c.f. reviewers' comments on,[58] e.g. Eric Bapteste is "agnostic" regarding the conclusions) and are often not mentioned in reviews (e.g.[59]) due to the subjective nature of the assumptions made.[60] However, despite there being a wealth of statistically supported studies towards the rooting of the tree of life between the Bacteria and the Neomura by means of a variety of methods,[61] including some that are impervious to accelerated evolution—which is claimed by Cavalier-Smith to be the source of the supposed fallacy in molecular methods[57]—there are a few studies which have drawn different conclusions, some of which place the root in the phylum Firmicutes with nested archaea.[62][63][64] Radhey Gupta's molecular taxonomy, based on conserved signature sequences of proteins, includes a monophyletic Gram negative clade, a monophyletic Gram positive clade, and a polyphyletic Archeota derived from Gram positives.[65][66][67] Hori and Osawa's molecular analysis indicated a link between Metabacteria (=Archeota) and eukaryotes.[68] The only cladistic analyses for bacteria based on classical evidence largely corroborate Gupta's results (see comprehensive mega-taxonomy). James Lake presented a 2 primary kingdom arrangement (Parkaryotae + eukaryotes and eocytes + Karyotae) and suggested a 5 primary kingdom scheme (Eukaryota, Eocyta, Methanobacteria, Halobacteria, and Eubacteria) based on ribosomal structure and a 4 primary kingdom scheme (Eukaryota, Eocyta, Methanobacteria, and Photocyta), bacteria being classified according to 3 major biochemical innovations: photosynthesis (Photocyta), methanogenesis (Methanobacteria), and sulfur respiration (Eocyta).[69][70][71] He has also discovered evidence that Gram-negative bacteria arose from a symbiosis between 2 Gram-positive bacteria.[72] AuthoritiesClassification is the grouping of organisms into progressively more inclusive groups based on phylogeny and phenotype, while nomenclature is the application of formal rules for naming organisms.[73] Nomenclature authorityDespite there being no official and complete classification of prokaryotes, the names (nomenclature) given to prokaryotes are regulated by the International Code of Nomenclature of Prokaryotes (Prokaryotic Code), a book which contains general considerations, principles, rules, and various notes, and advises[74] in a similar fashion to the nomenclature codes of other groups. Classification authoritiesAs taxa proliferated, computer aided taxonomic systems were developed. Early non networked identification software entering widespread use was produced by Edwards 1978, Kellogg 1979, Schindler, Duben, and Lysenko 1979, Beers and Lockhard 1962, Gyllenberg 1965, Holmes and Hill 1985, Lapage et al 1970 and Lapage et al 1973.[75]: 63 Today the taxa which have been correctly described are reviewed in Bergey's manual of Systematic Bacteriology, which aims to aid in the identification of species and is considered the highest authority.[42] An online version of the taxonomic outline of bacteria and archaea (TOBA) is available [1]. List of Prokaryotic names with Standing in Nomenclature (LPSN) is an online database based on the International Code of Nomenclature of Prokaryotes which currently contains over two thousand accepted names with their references, etymologies and various notes.[76] Description of new speciesThe International Journal of Systematic Bacteriology/International Journal of Systematic and Evolutionary Microbiology (IJSB/IJSEM) is a peer reviewed journal which acts as the official international forum for the publication of new prokaryotic taxa. If a species is published in a different peer review journal, the author can submit a request to IJSEM with the appropriate description, which if correct, the new species will be featured in the Validation List of IJSEM. DistributionMicrobial culture collections are depositories of strains which aim to safeguard them and to distribute them. The main ones being:[73]
Alternative systemsA few other nomenclatural systems have been proposed to correct for perceived shortcomings in the Prokaryotic Code system:
These following systems provide a taxonomy database under more ad hoc rules:
AnalysesBacteria were at first classified based solely on their shape (vibrio, bacillus, coccus etc.), presence of endospores, gram stain, aerobic conditions and motility. This system changed with the study of metabolic phenotypes, where metabolic characteristics were used.[83] Recently, with the advent of molecular phylogeny, several genes are used to identify species, the most important of which is the 16S rRNA gene, followed by 23S, ITS region, gyrB and others to confirm a better resolution. The quickest way to identify to match an isolated strain to a species or genus today is done by amplifying its 16S gene with universal primers and sequence the 1.4kb amplicon and submit it to a specialised web-based identification database, namely either Ribosomal Database Project[2] Archived 19 August 2020 at the Wayback Machine, which align the sequence to other 16S sequences using infernal, a secondary structure bases global alignment,[84][85] or ARB SILVA, which aligns sequences via SINA (SILVA incremental aligner), which does a local alignment of a seed and extends it [3].[86] Several identification methods exists:[73]
New speciesThe minimal standards for describing a new species depend on which group the species belongs to. c.f.[87] CandidatusCandidatus is a component of the taxonomic name for a bacterium that cannot be maintained in a Bacteriology Culture Collection. It is an interim taxonomic status for noncultivable organisms. e.g. "Candidatus Pelagibacter ubique" Species conceptBacteria divide asexually and for the most part do not show regionalisms ("Everything is everywhere"), therefore the concept of species, which works best for animals, becomes entirely a matter of judgment. The number of named species of bacteria and archaea (approximately 21,000)[88] is surprisingly small considering their early evolution, genetic diversity and residence in all ecosystems. The reason for this is the differences in species concepts between the bacteria and macro-organisms, the difficulties in growing/characterising in pure culture (a prerequisite to naming new species, vide supra) and extensive horizontal gene transfer blurring the distinction of species.[89] The most commonly accepted definition is the polyphasic species definition, which takes into account both phenotypic and genetic differences.[90] However, a quicker diagnostic ad hoc method to use a purely genetic approach, including any one of:
It has been noted that if the 70% DDH threshold were applied to animal classification, the order primates would be a single species.[100] For this reason, more stringent species definitions based on whole genome sequences have been proposed. Specifically, Wright et al. (2018) goes beyond ANI and AF to propose defining species as a group in which the maximum distance between any two members is greater than the minimum distance with any outsider. This criterion can be put on top of ANI+AF without introducing too many splits.[97] Pathology vs. phylogenyIdeally, taxonomic classification should reflect the evolutionary history of the taxa, i.e. the phylogeny. Although some exceptions are present when the phenotype differs amongst the group, especially from a medical standpoint. Some examples of problematic classifications follow. Escherichia coli: overly large and polyphyleticIn the family Enterobacteriaceae of the class Gammaproteobacteria, the species in the genus Shigella (S. dysenteriae, S. flexneri, S. boydii, S. sonnei) from an evolutionary point of view are strains of the species Escherichia coli (polyphyletic), but due to genetic differences, cause different medical conditions in the case of the pathogenic strains.[101] Confusingly, there are also E. coli strains that produce Shiga toxin known as STEC. The new average nucleotide identity (ANI) criterion, as used by GTDB, groups most samples (including Shigella) into one species, but spreads the rest in five species. A definition using the biological species concept (specifically, presence of recombination) found that all but 12 genomes tagged as E. coli in GenBank fall into one "species", with the lowest strain-to-strain ANI being 94%.[102] Bacillus cereus group: close and polyphyleticIn a similar way, the Bacillus species (=phylum Firmicutes) belonging to the "B. cereus group" (B. anthracis, B. cereus, B . thuringiensis, B. mycoides, B. pseudomycoides, B. weihenstephanensis and B. medusa) have 99-100% similar 16S rRNA sequence (97% is a commonly cited adequate species cut-off) and are polyphyletic, but for medical reasons (anthrax etc.) remain separate.[103] The new ANI criterion provides some support for members of this group as separate species.[104] Yersinia pestis: extremely recent speciesYersinia pestis is in effect a strain of Yersinia pseudotuberculosis, but with a pathogenicity island that confers a drastically different pathology (Black plague and tuberculosis-like symptoms respectively) which arose 15,000 to 20,000 years ago.[105] Nested genera in PseudomonasIn the gammaproteobacterial order Pseudomonadales, the genus Azotobacter and the species Azomonas macrocytogenes are actually members of the genus Pseudomonas, but were misclassified due to nitrogen fixing capabilities and the large size of the genus Pseudomonas which renders classification problematic.[83][106][107] This will probably rectified in the close future. Nested genera in BacillusAnother example of a large genus with nested genera is the genus Bacillus, in which the genera Paenibacillus and Brevibacillus are nested clades.[108] By 2020 there was enough genomic data to resolve Bacillus and split out 23 genera. However, even this was insufficient to make Bacillus monophyletic, because microbiologists do not want to move the pathogenic Bacillus cereus group out of the genus yet. B. cereus is separated from the type species B. subtilis by many genus-sized clades, many of which have been made into formal genera.[109][110] Agrobacterium: resistance to name changeBased on molecular data it was shown that the genus Agrobacterium is nested in Rhizobium and the Agrobacterium species transferred to the genus Rhizobium (resulting in the following comp. nov.: Rhizobium radiobacter (formerly known as A. tumefaciens), R. rhizogenes, R. rubi, R. undicola and R. vitis)[111] Given the plant pathogenic nature of Agrobacterium species, it was proposed to maintain the genus Agrobacterium[112] and the latter was counter-argued.[113] The problem was resolved in the 2010s by the reinstatement of Agrobacterium via splitting genera, after Rhizobium was also split a few times.[114][115] The genus Agrobacterium is to only include the clade clustered around the type species Agrobacterium radiobacter, with a clear synapomorphy in the form of the protelomerase telA gene.[116][117] Other examples of name-change resistance
|